Fantastic Tips About How To Detect Copy Number Variation

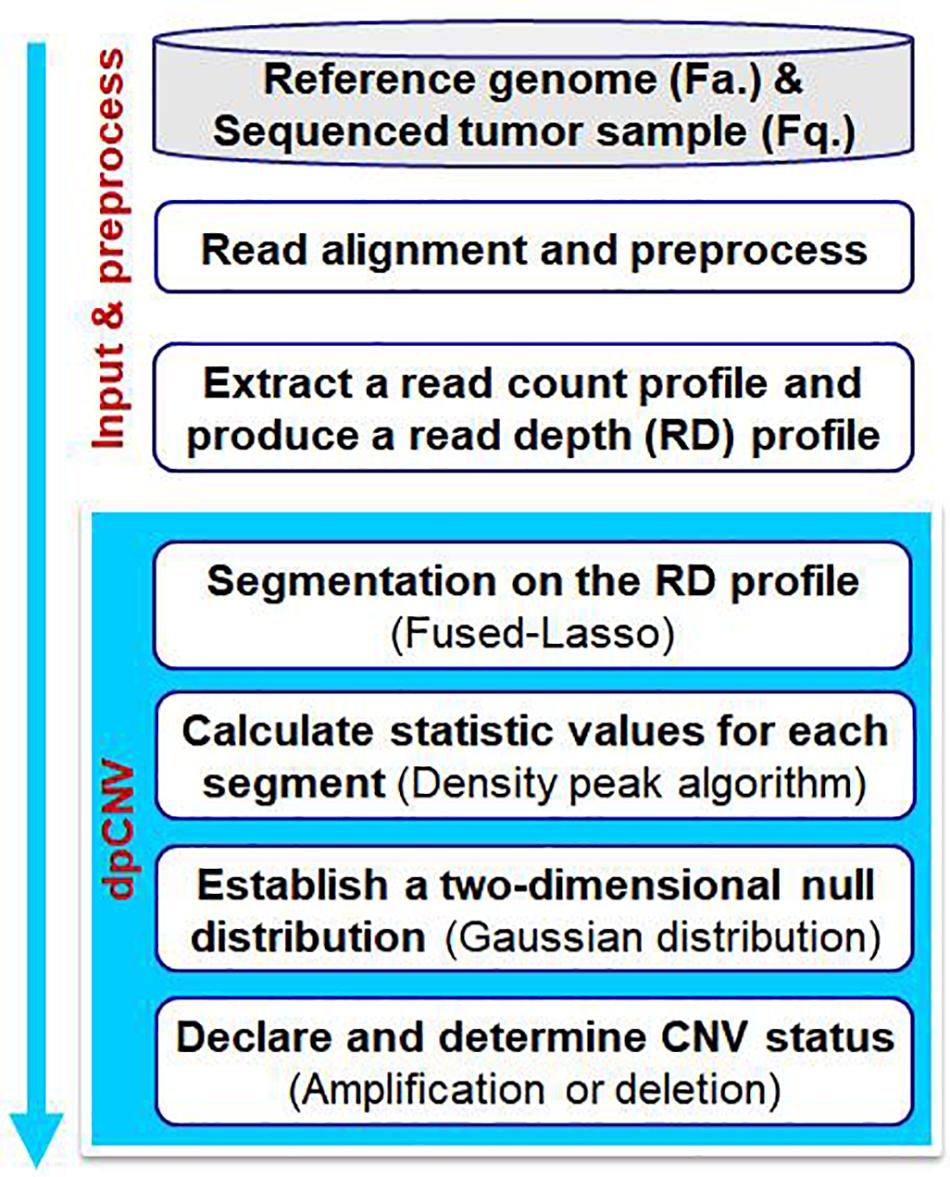
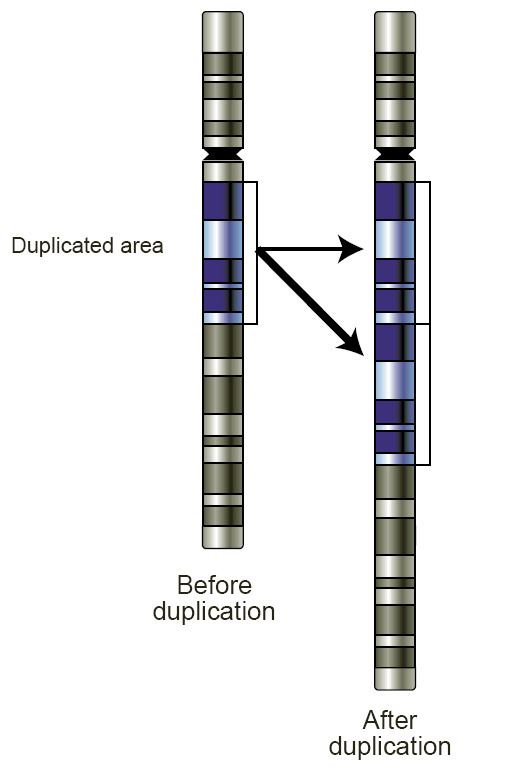
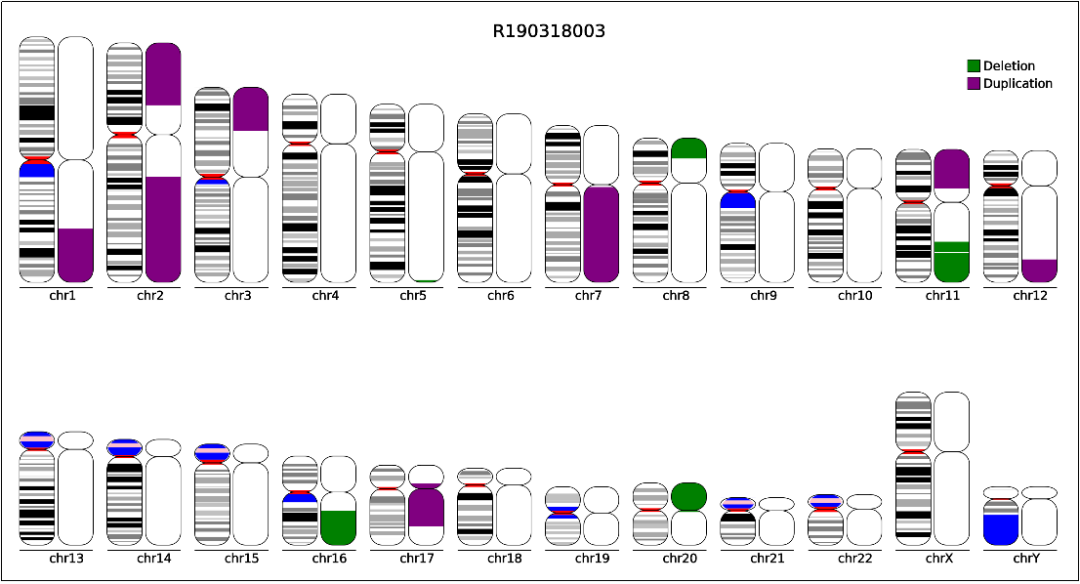
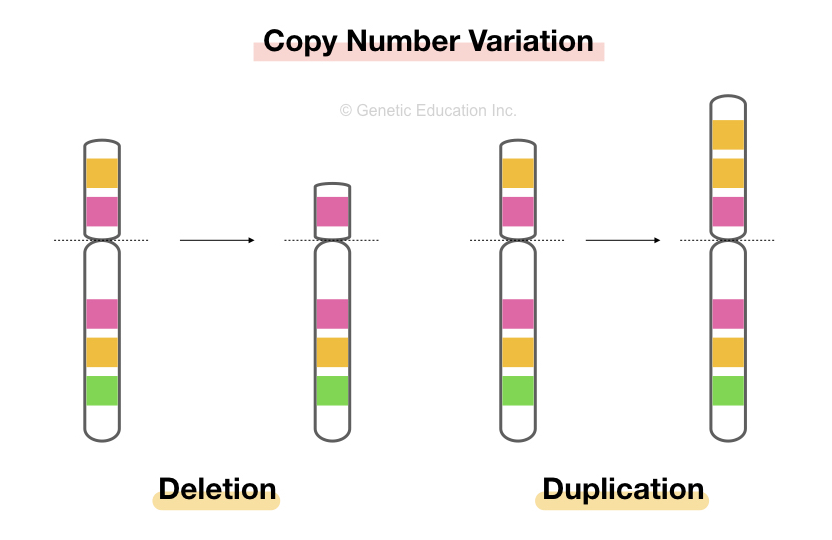
Copy number aberrations (cnas), which are pathogenic copy number variations (cnvs), play an important role in the initiation and progression of cancer.
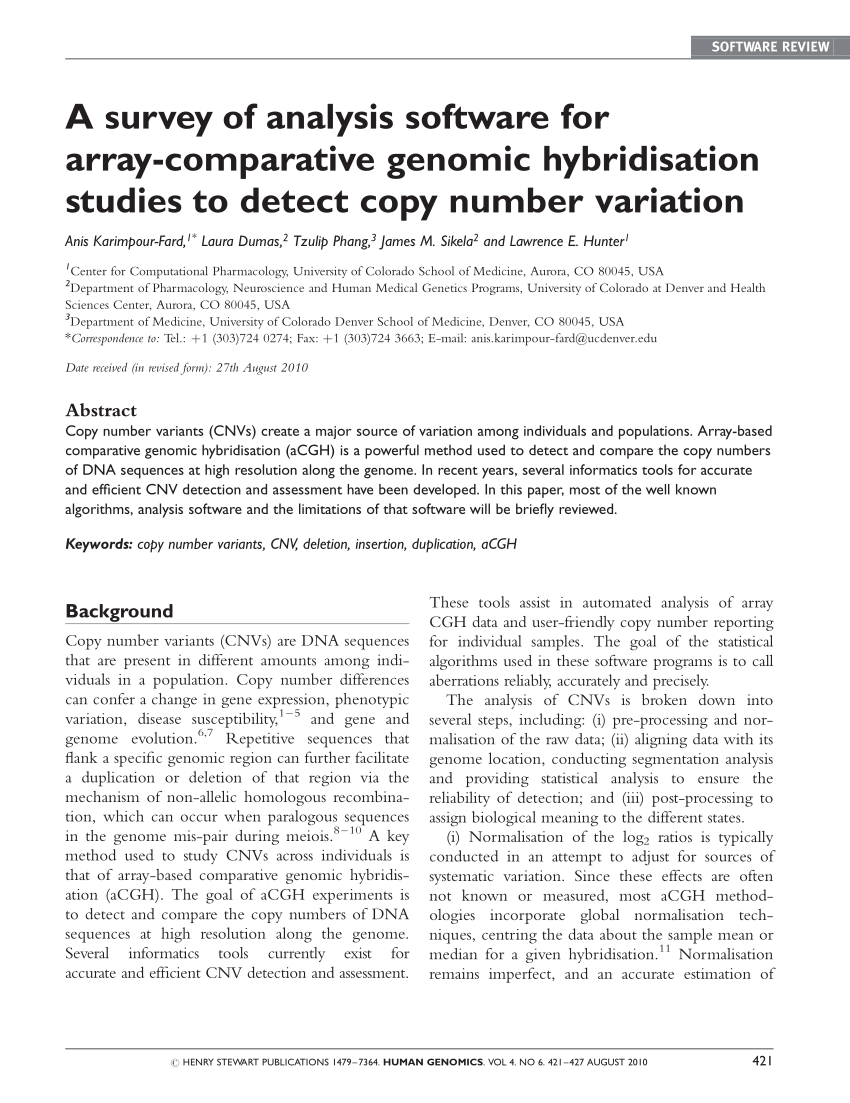
How to detect copy number variation. Copy number variants (cnv) are associated with phenotypic variation in several species. However, there are no clear boundaries between the two. Recently copy number variation (cnv) has gained considerable interest as a type of genomic/genetic variation that plays an important role in disease susceptibility.
[1] copy number variations can be generally categorized into two main groups: Cgh was first developed as a. Integration of multiple genetic sources for copy number variation detection (cnv) is a powerful approach to improve the identification of variants.
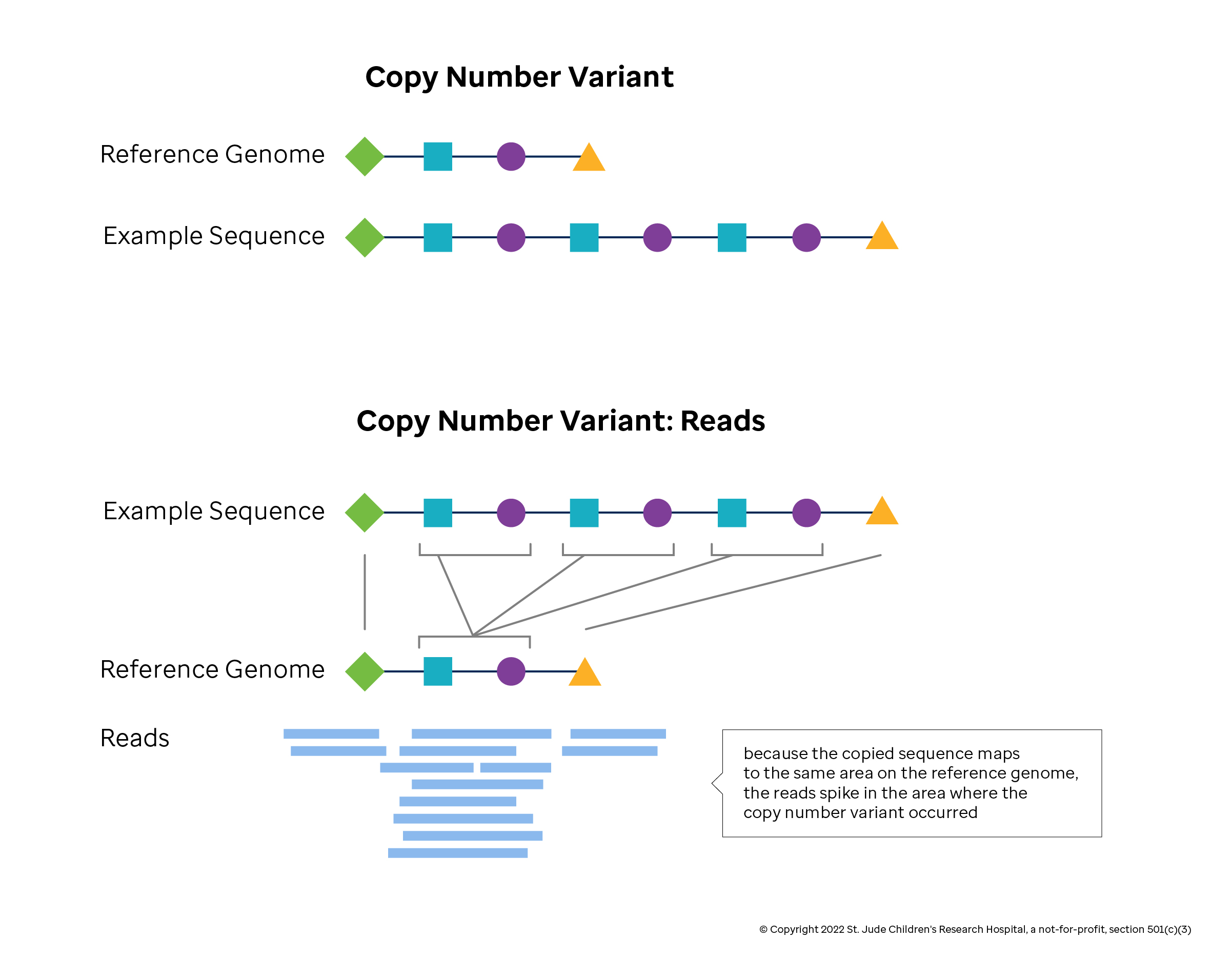
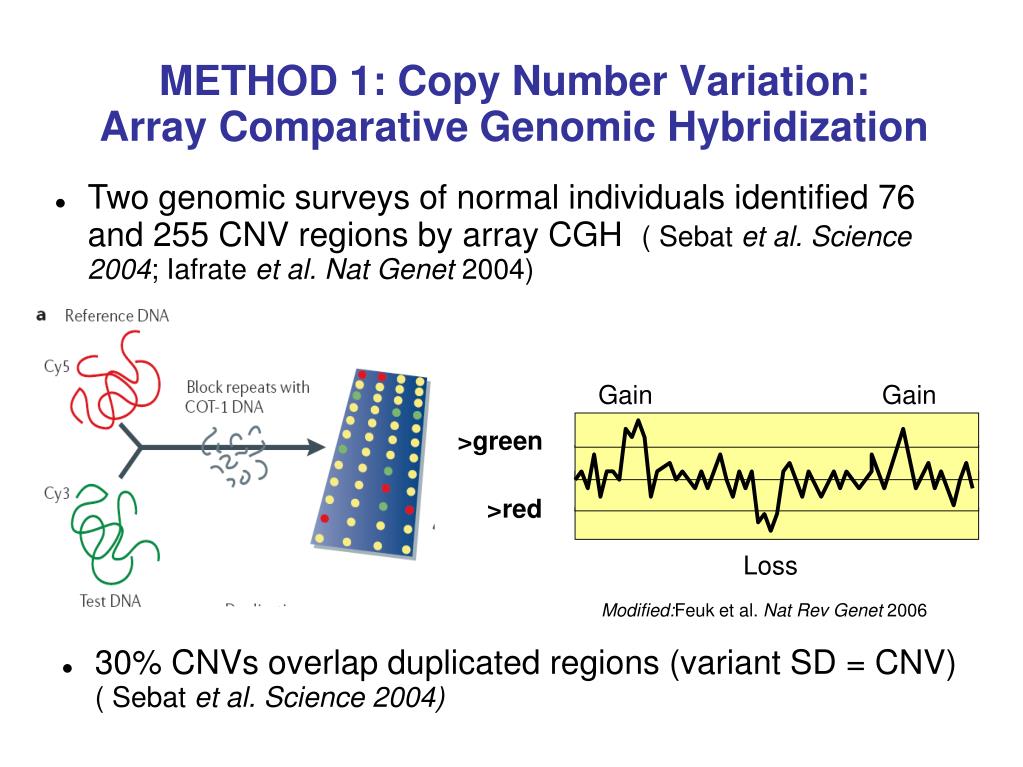
One of the main methods by which cnvs can be identified using dna microarrays is comparative genomic hybridization (cgh). For detecting deletions, 95% power is achieved for. Author summary as an important type of genomic structural variation, cnvs are associated with complex phenotypes because they change the.
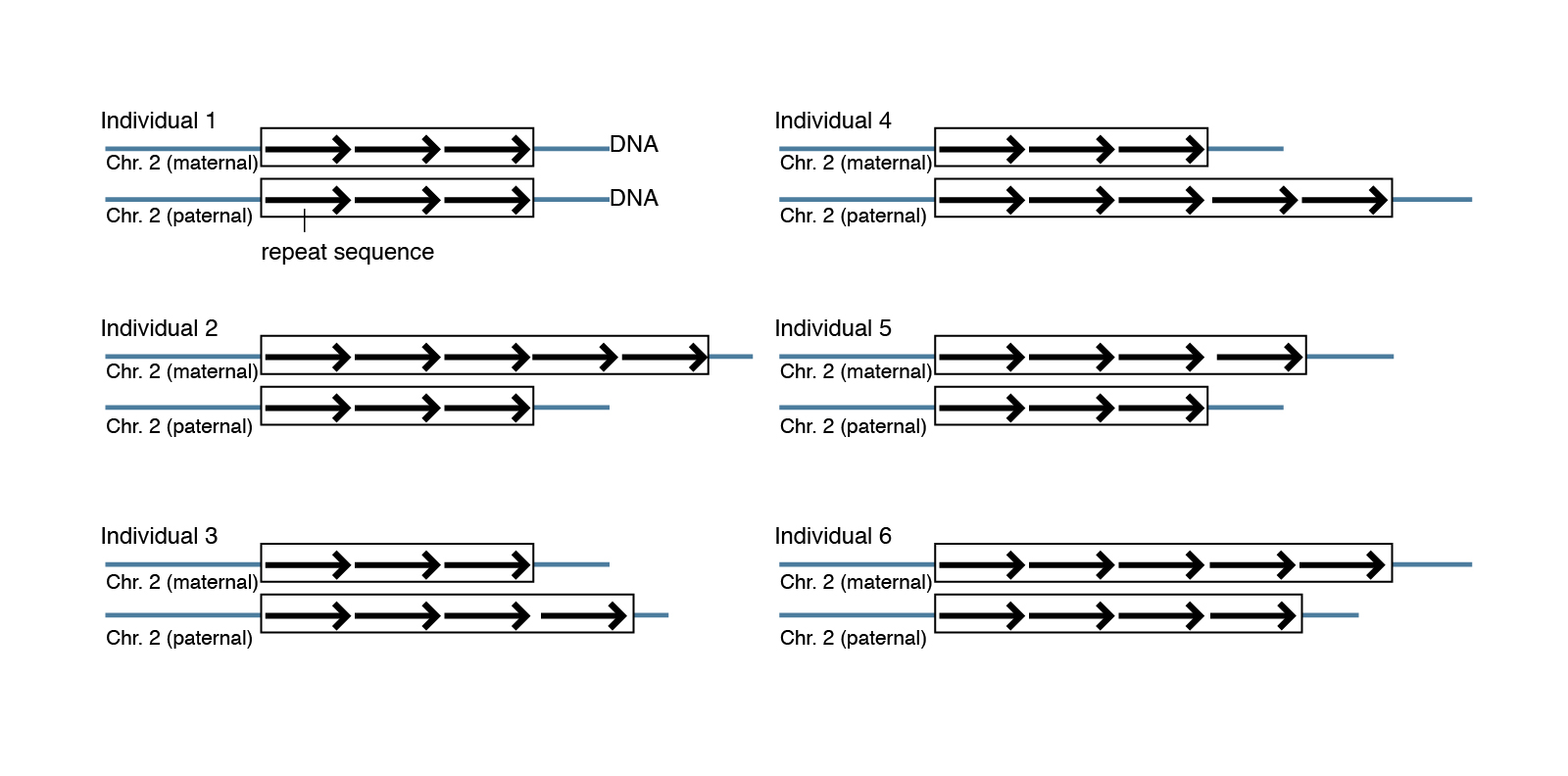
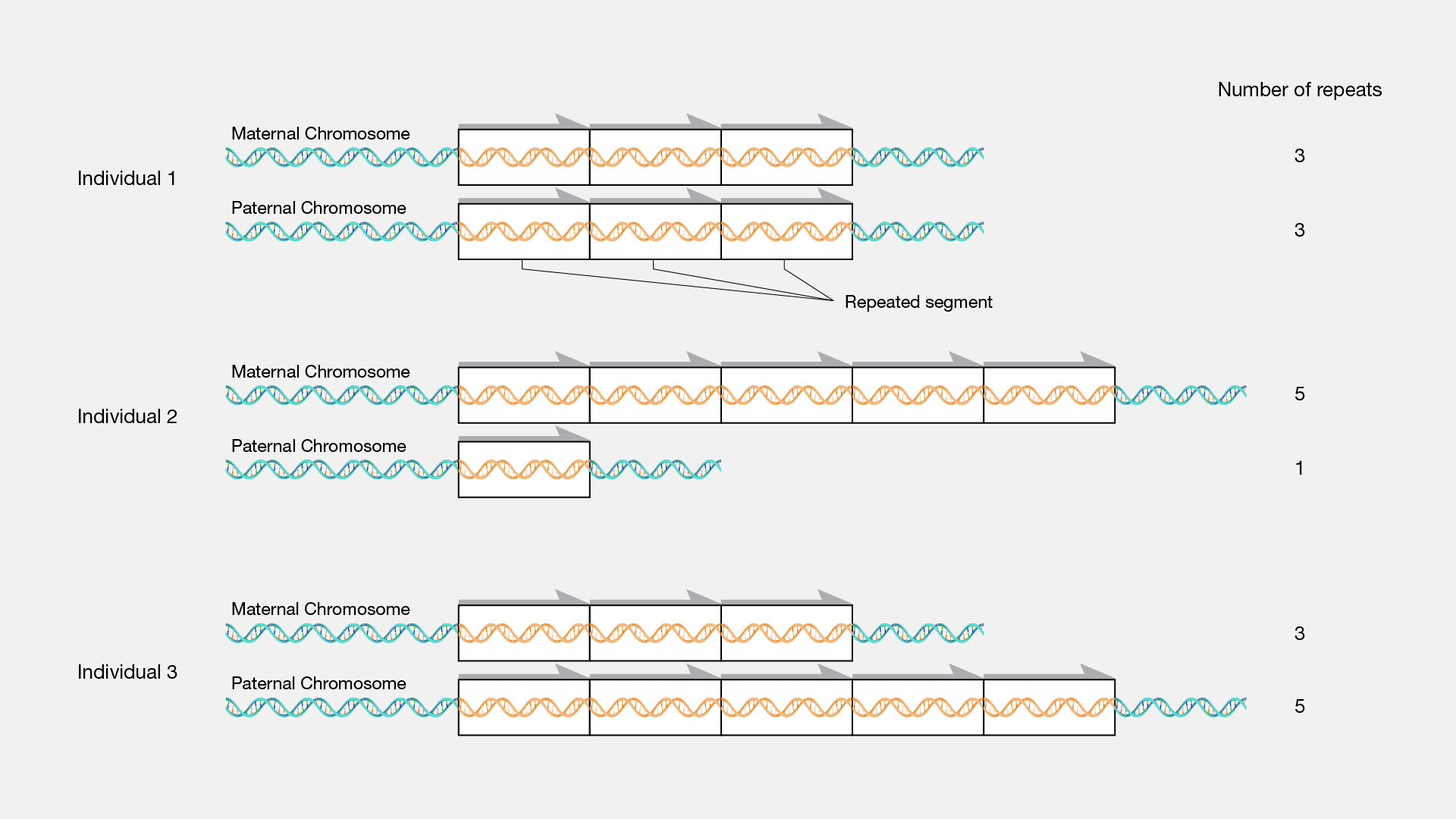
One of the most traditional methods to evaluate the copy number variations visually is the karyotyping. Trost b, walker s, wang z, thiruvahindrapuram b, macdonald jr, sung wwl, et al. Short repeats and long repeats.
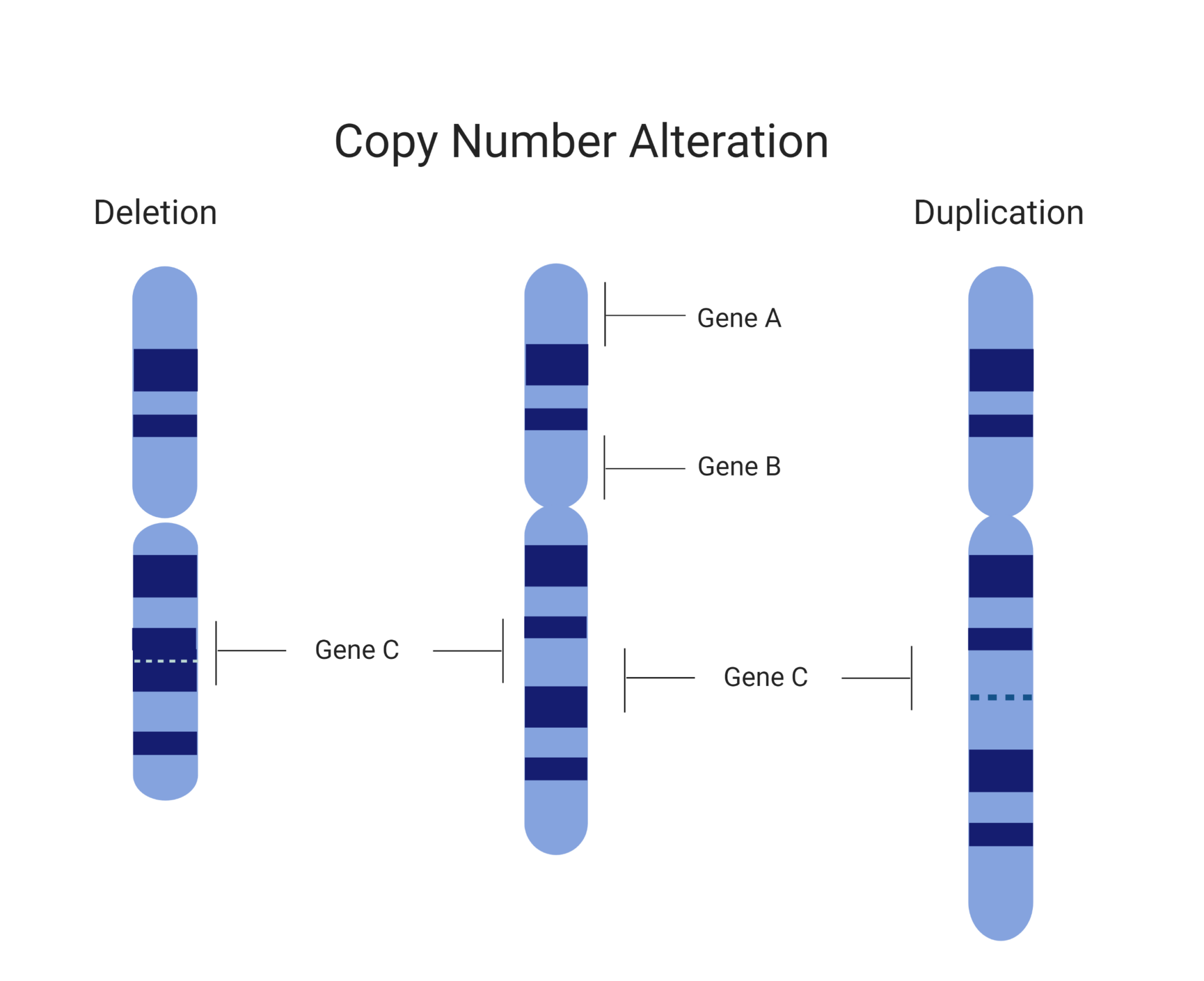
Comparative genomic hybridization to arrays one of the main methods by which cnvs can be identified using dna microarrays is comparative genomic hybridization. Using a maximum of likelihood ratio statistics to detect narrower segments of aberration, cbs ( 64) was able to divide the genome into regions of equal dna copy. However, properly detecting changes in copy numbers of sequences.
Techniques to detect copy number variations: Resolution of cnv detection with exomecnv is limited largely by the probe design: